Epigenetic modifications, such as DNA methylation and histone modification, are one of the most important factors in the regulation of gene expression. Differential gene expression caused by DNA methylation plays an important role in cancer. For example, cancer cells often have hypomethylation of oncogenes and hypermethylation of tumor suppressor genes, but the mechanisms in the regulation of DNA methylation are not fully understood. In addition, recent researches showed that RNA and RNA modifications may play regulatory roles in gene expression and cancer as well. The most abundant RNA modification is N6-Methyladenosine (m6A), which takes part in various biological process, such as RNA stability and translation efficiency. Recent studies have also shown that RNA-m6A modification can reversely regulate chromatin remodeling and histone modification. However, the relationship between RNA m6A and DNA methylation is still undescribed.
On September 8, 2022, a new study titled RNA m6A regulates transcription via DNA demethylation and chromatin accessibility, jointly conducted by Prof. Lin Dongxin/ Prof. Zheng Jian laboratory from Sun Yat-sen University Cancer Center and Prof. Chen Jianjun from Beckman Institute, City of Hope National Medical Center, USA was published online in Nature Genetics (Figure 1). This study found that RNA m6A modification directly demethylates adjacent DNA 5mC co-transcriptionally, resulting in alteration of chromatin accessibility and gene expression. This study revealed for the first time that RNA m6A can regulate DNA methylation, which is of great significance for further understanding the complex regulatory mechanism of gene expression.

Figure 1: The research published in Nature Genetics
In this issue, researchers showed that the RNA m6A reader FXR1 recruits DNA 5-methylcytosine dioxygenase TET1 to proximal genomic regions co-transcriptionally, resulting in DNA demethylation (Figure 1). To explore the biological function of RNA m6A regulating DNA demethylation, they further investigated deeply in esophageal squamous cancer. They found loss of METTL3, TET1 or FXR1 could lead to changes in chromatin accessibility of localized regions, and then affect the transcription of corresponding genes. RNA m6A in esophageal squamous cancer tissues was significantly higher than that in adjacent normal tissues, but DNA methylation levels were decreased. Knockdown of METTL3, TET1 or FXR1 can inhibit the proliferation, invasion and migration of esophageal squamous cancer cells, and FXR1 function depends on the existence of METTL3 and TET1, further indicating that RNA m6A negatively regulates DNA methylation and plays an important role in the esophageal squamous cancer.
Deng Shuang, Zhang Jialiang, Su Jiachun,and Zuo Zhizhi are the co-first authors of this paper. Prof. Zheng Jian, Prof. Lin Dongxin and Prof. Chen Jianjun are the co-corresponding authors of the research paper.
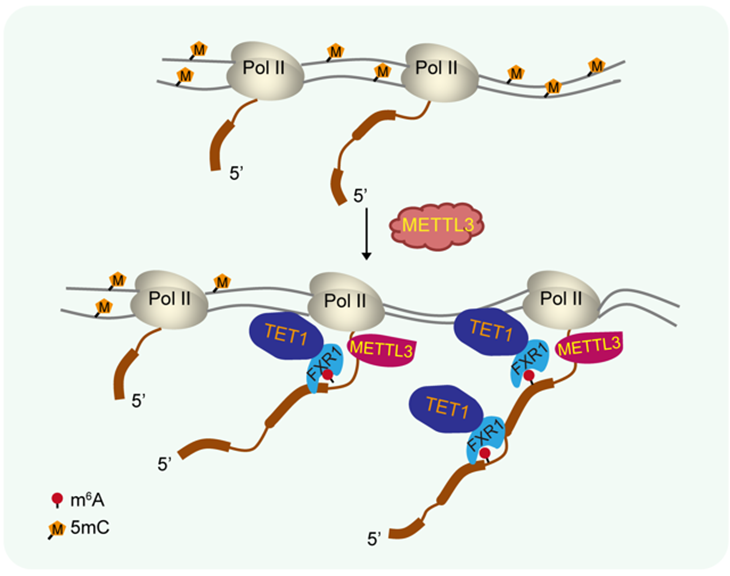
Figure 2. RNA-m6A recruits TET1 through FXR1 to remove DNA methylation
In the meanwhile, Prof. Shen Hongjie from the Institute of Biomedical Sciences, Fudan University, was invited by Nature Genetics to publish a news & views titled When RNA methylation meets DNA methylation. This article highly evaluates the mechanism that RNA m6A recruits TET1 through m6A reader FXR1 to regulate DNA methylation for the first time, and confirms the important role of this mechanism in the regulation of chromatin accessibility, gene expression and esophageal squamous cancer. They considered this study might provide a new direction for further research on the treatment of ESCC and regulation of epigenetic modifications in the future.
The Laboratory of Academician Lin Dongxin of Sun Yat-sen University Cancer Center established in 2015, they focus on genomics and epigenetics in cancer. The team recruits post doctors and researchers, qualified candidates with background in oncology, biochemistry, molecular biology and bioinformatics are warmly welcomed. (Contact:zhengjian@sysucc.org.cn).
Copyright:Sun Yat-sen University Cancer Center Designed by Wanhu.